|
| |
|
|
|
|
The BxDesignertm is used in the experimental
design process to create custom microarrays and to manage probe data. The descriptions
of the different types of DNA and Protein microarrays is
focused on presenting an architectural view for
designing relational databases to handle each type. The
template database schemas are designed to hold the fundamental
information about microarrays and they are linked to other
tables that contain information about
the manufactured or created microarrays. For example, the additional tables
may contain the date of production, lot
and part numbers, company, expiration date (if
applicable), operator, chemical component sources and unique microarray IDs.
Probes
DNA Probes are cDNA or Oligos that have a length of about 25 nucleotide
bases. Other types of Probes are proteins, antibodies, ligands or other types
of molecules that will bind probes. BxLibrariantm will manage each probe using a
unique ID. For example, information about the source of the probe, its well
location within the microtitre plate, its microtitre plate ID, its sequence and probe handling protocols will be
linked to the probe's unique ID.
Slides/Chips
Slides and/or Chips have bound targets arranged in patterns. These patterns
are determined by the type of experiments to be performed and by the type of
comparative or differential analysis that is to be done. One type of strategy is
to be able to have self validating and internal control targets. Self-validating
means that duplicate targets are present and if one target is expressed than the
other has to be expressed if the first is not to be considered a false
expression. Internal control targets allow for normalizing the data in order to
compare expressions from slide to slide. The database must have a mechanism for
storing the pattern and relationships between targets within and across patterns
and to know the different types of targets. It must also store the protocols for
creating the slides or chips. Therefore, each slide will have a unique ID
and may also have lot or batch numbers.
NOTE: The Microsoft SQL Server 2000 database
schemas that are presented should be viewed as a base schema upon
which to build or extend. They may not be complete due
to specifics of a particular microarray and the needs of the downstream instrument control and data mining
applications. They also can be implemented on other client-server databases
such as Oracle, Sybase and DB2. |

|

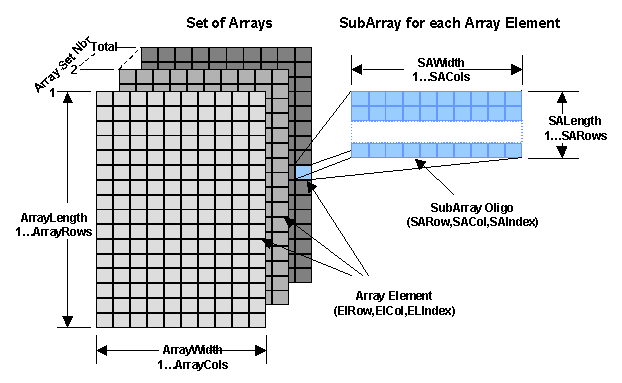
| The following figure depicts an architecture
of a set of oligo microarrays. Each microarray has a set
number that ranges from 1 to the total number of arrays in the set.
In addition each microarray has a length (ArrayLength), a width (ArrayWidth)
and a number of rows (ArrayRows) and columns (ArrayCols).
An array element is accessed by a row (ElRow)
and column (ElCol) coordinate or an equivalent index (ELIndex)
that runs from 1 to the total number (ArrayRows*ArrayCols) of elements
in the array. The element index is calculated
by (ElRow-1)*ArrayCols + ElCol. Each element is composed of
a Subarray of oligo sequences where each is different from any other
oligo in the array. The Subarray has a widh (SAWidth) and a length
(SALength) and a number of columns (SACols) and rows (SARows). Each Oligo is
accessed by a row (SARow) and column (SACol) coordinate or an equivalent index
(SAIndex) that runs from 1 to the total number of oligos
(SARows*SACols).
|


Top |

|
This database schema stores the information about the Oligo
Microarray Set described above and the term template is used
to designate that many production microarrays will be manufactured
based upon this information. In addition to the physical
attributes of the microarray set, dates, versions, descriptions of
each array, species and reference information about each oligo
is stored. The coordinates in the scanned image of the expressed
values will be mapped to the coordinates in the template.
Additional database tables will be needed to
store manufacturing information such as part numbers, lot
numbers, date of manufacture and other related data.
Each microarray will be assigned a unique ID for tracking back
to the manufacturing process and forward through
the experimental process and analysis.
 |


Top |

|
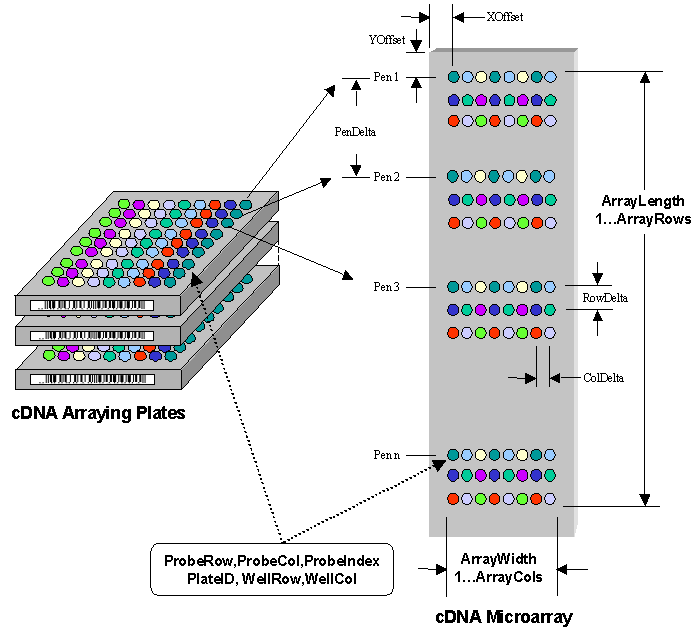
The following figure depicts three
major components of a cDNA system. A set of arraying plates containing
the cDNA, a linear set of Pens
(not shown) that extract cDNA from the wells of the arraying plates and array the
cDNA in a pattern onto the third component, which is a microarray with a prepared surface,
that binds the cDNA. Each arraying plate has a unique ID (PlateID) and each
well has a row (WellRow) and a column (WellCol)
coordinate. A linear pen array transfers the cDNA from the plates to the microarray with
the distance (PenDelta) from one pen to another being the same as
the distance from one well to another. There are a number of specified position values -
an upper left corner of the array (XOffset, YOffset) and the
distance between rows (RowDelta) and columns (ColDelta). The microarray has a length (ArrayLength), a width (ArrayWidth)
and a number of rows (ArrayRows) and columns (ArrayCols). Each Probe has
a diameter that ranges from 50 to 300 microns
and a row (ProbeRow) and column (ProbeCol) coordinate or
an equivalent index (ProbeIndex) that runs from 1 to the total number of Probes (ArrayRows*ArrayRows) on
the array. It is also linked back to the well in the
source microtitre plate. In addition, complex patterns can be created that
may consist of more than a single deposition of the cDNA and consist
of controls.
|


Top
|

|
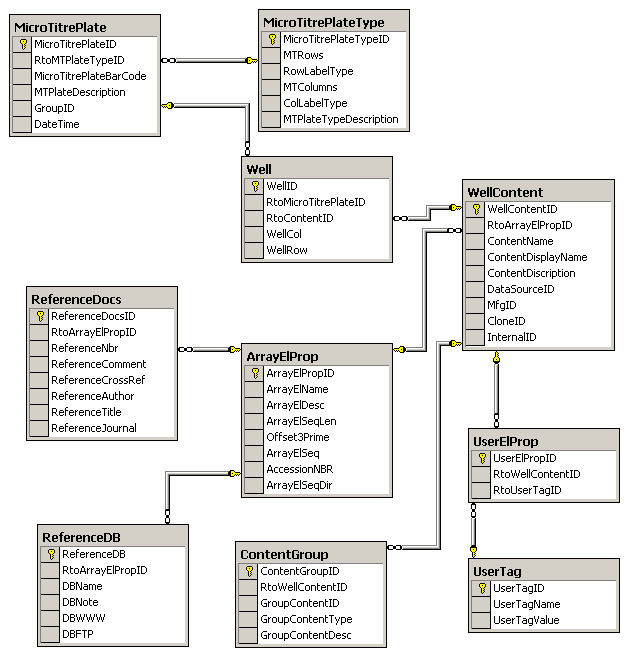
The first part of the database schema
handles the data related to the microtitre arraying plates. The
data contains information about the contents of each well which
includes links to multiple databases and reference
articles. It also provides for user defined properties where
the user can supply a name and a value for the property. The
user defined properties are applied uniquely to each WellContentID and thus disparate information can be
stored.
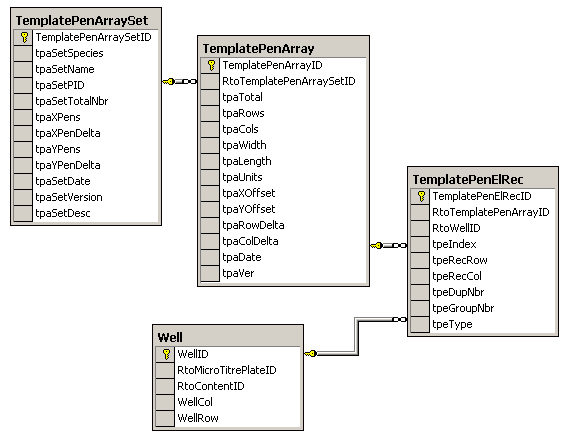
This part of the schema handles the
pen layout data for the arrayed microarrays and shows the link to the first part
that handles the microtitre arraying plates through the Well table. This information
is directly related to the expressed values in the scanned images. In general these tables can be
extended to utilize a dynamic arraying pattern model.
|


Top
|

|
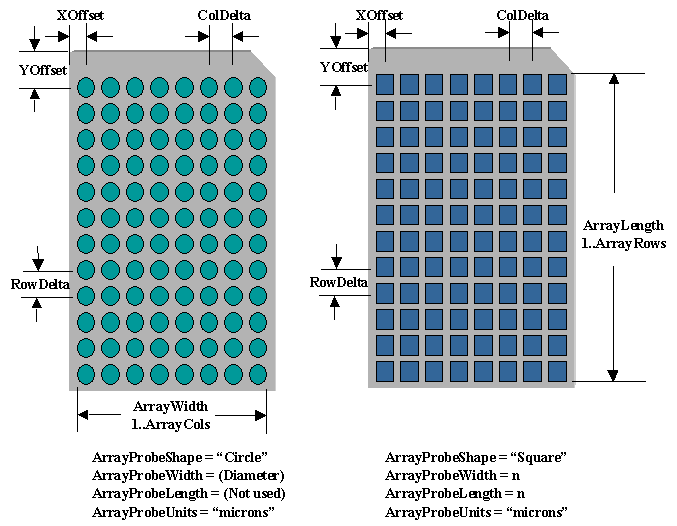
The following figure shows two different protein arrays where
the affinity capture surfaces are arrayed in a standard
microtitre plate arrangement. In this example an 8 x12 array;
however, other standard microtitre
plate sizes (e.g. 384, 1536) and non-standard sizes (10,000 locations)
can be created. In addition, the shape of
the surface can be a circle or any
polygon and the surface does not have to be homogeneous and all
surfaces can be different from each other. The
database schema stores the array pattern, shapes
and properties of the affinity capture surface. The affinity capture
surface can be antibodies, antigens, proteins and other
specially prepared chemical surfaces that differentially bind molecules such as proteins.
|


Top
|

|
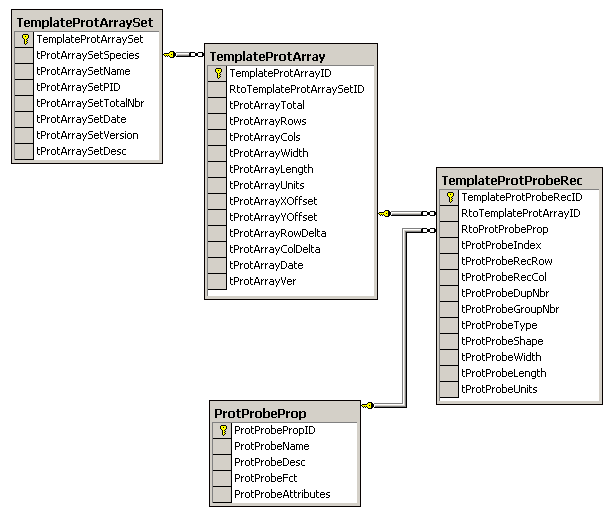
The schema for Protein Probe Arrays supports the Protein
Array figure above and is similar to the previous DNA
microarrays. The fields in the TemplateProtProbeRec table that
pertain to the shape of the affinity capture surface allows for each
site on the array to have a different shape. These fields could be
moved to the TemplateProtArray table if all shapes on the array are
the same. Each protein array will have its own instance of the set
of Template Protein Tables. The detection system can be controlled
by this information, especially the shape data, as well as
being linked to the results of the anlysis which can be a
fluorescent image or a mass(m/z) vs. Intensity
spectrum. The properties of the surfaces are important for
helping in the identification of proteins that are captured;
therefore, the ProtProbeProp table can be extended with more fields
to contain more information about the surface.
|

Top |
| |
